Cardiac LV 17-segment model#
This notebook demonstrates how to use the geometric algorithm to automatically segment the 17-segment Left Ventricle myocardium model defined by the AHA: https://www.ahajournals.org/doi/pdf/10.1161/hc0402.102975
[1]:
try:
import platipy
except:
# Install platipy with the 'cardiac' extra since that contains some extra libraries we need.
!pip install platipy[cardiac]
import platipy
from pathlib import Path
import matplotlib.pyplot as plt
import SimpleITK as sitk
import numpy as np
from platipy.imaging import ImageVisualiser
from platipy.imaging.label.utils import get_com
from platipy.imaging.utils.io import write_nrrd_structure_set
from platipy.imaging.utils.ventricle import generate_left_ventricle_segments
from platipy.imaging.projects.cardiac.run import install_open_atlas
Download Sample Data#
PlatiPy’s cardiac segmentation tool uses an atlas as part of that segmentation. For this example we download that atlas to demonstrate the 17-segment auto-segmentation.
[2]:
atlas_path = Path("data/atlas")
if not atlas_path.exists():
install_open_atlas(atlas_path)
Read Data#
We read in some of the data we downloaded. The required structures are: Left Ventricle, Left Atrium, Right Ventricle and the whole heart. The CT image is loaded but is only needed for visualisation purposes.
[3]:
patid = "LUNG1-002"
image_path = atlas_path.joinpath(patid, "IMAGES", "CT.nii.gz")
image = sitk.ReadImage(str(image_path)) # onyl used for visualisation
contours = {}
lv_path = atlas_path.joinpath(patid, "STRUCTURES", "Ventricle_L.nii.gz")
contours["Ventricle_L"] = sitk.ReadImage(str(lv_path))
la_path = atlas_path.joinpath(patid, "STRUCTURES", "Atrium_L.nii.gz")
contours["Atrium_L"] = sitk.ReadImage(str(la_path))
rv_path = atlas_path.joinpath(patid, "STRUCTURES", "Ventricle_R.nii.gz")
contours["Ventricle_R"] = sitk.ReadImage(str(rv_path))
heart_path = atlas_path.joinpath(patid, "STRUCTURES", "Heart.nii.gz")
contours["Heart"] = sitk.ReadImage(str(heart_path))
Generate 17 LV Segments#
The generate_left_ventricle_segments function is run which returns a dictionary where the key is the segment ID (i.e. Ventricle_L_Segment1, Ventricle_L_Segment2, …) and the value is the segment auto-contour (as a SimpleITK.Image).
See the documentation for additional options you can specify.
[4]:
lv_segments = generate_left_ventricle_segments(contours, verbose=True)
Beginning LV segmentation algorithm.
Module 1: Cropping and initial alignment.
Images cropped. Volume reduction: 1.000
Alignment computed.
Cardiac axis: [-0.51538678 -0.8567381 0.01939842]
Rotation axis: [-0.8567381 -0.01939842 0. ]
Rotation angle: 1.029337402528849
Rotation centre: (53.54705757769439, -33.01797661455572, -33.16549199516518)
Module 2: LV orientation alignment.
Optimiser tolerance (degrees) = 1
Beginning alignment process
N: 1
LV apex: [ 92.12689862 -29.34676304 -100.3999939 ]
MV COM: [ 38.60792831 -34.98956324 -14.50176381]
LV axis: [ 53.51897031 5.6428002 -85.89823009]
Rotation axis: [ 5.6428002 -53.51897031 0. ]
Rotation centre: [ 65.36741347 -32.16816314 -57.45087885]
Rotation angle: 0.5596804399185753
N: 2
LV apex: [ 74.15895319 -25.37873232 -112.3999939 ]
MV COM: [ 65.39114152 -32.17008994 -6.80293245]
LV axis: [ 8.76781167 6.79135762 -105.59706144]
Rotation axis: [ 6.79135762 -8.76781167 0. ]
Rotation centre: [ 69.77504736 -28.77441113 -59.60146318]
Rotation angle: 0.10464206469597664
N: 3
LV apex: [ 71.08173551 -27.39587178 -112.3999939 ]
MV COM: [ 69.79644172 -28.76506973 -6.63819614]
LV axis: [ 1.28529379 1.36919795 -105.76179775]
Rotation axis: [ 1.36919795 -1.28529379 0. ]
Rotation centre: [ 70.43908861 -28.08047075 -59.51909502]
Rotation angle: 0.01775451288291543
N: 4
LV apex: [ 70.10473551 -28.37287177 -112.3999939 ]
MV COM: [ 70.77344171 -27.78806973 -6.63819614]
LV axis: [ -0.6687062 -0.58480204 -105.76179775]
Rotation axis: [-0.58480204 0.6687062 0. ]
Rotation centre: [ 70.43908861 -28.08047075 -59.51909502]
Rotation angle: 0.008399315304762168
Module 3: Myocardium generation.
Apex (long axis) slice: 9
Apical section extent slice: 19
Mid section extent slice: 29
Basal section extent slice: 40
DeltaCut (DC): 10
Extent: 31
Module 4: Segment generation.
RV basal slices: [29 30 31 32 33]
RV insertion angle (basal section): 1.5480729659532555
Apical LV-RV COM angle: 3.04084204317622
Computing apical segments
Computing mid segments
Computing basal segments
Module 5: Re-orientation.
Complete!
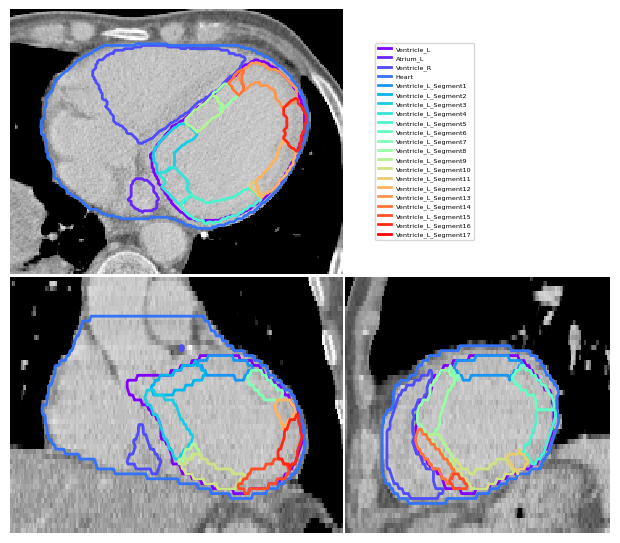
Visualise Segments#
Here we generate a figure using the CT loaded to visualise the results.
[5]:
vis = ImageVisualiser(image, cut=get_com(contours["Ventricle_L"]), figure_size_in=6)
vis.add_contour(contours)
vis.add_contour(lv_segments)
vis.set_limits_from_label(contours["Heart"], expansion=20)
fig = vis.show()
Optionally save a NRRD file containing all LV segments. This is useful for loading into Slicer (for example).
[6]:
write_nrrd_structure_set(lv_segments, atlas_path.joinpath(patid, "STRUCTURES", "LV_Segments.nrrd"), colormap=plt.cm.rainbow)
[ ]: