Contour Comparison#
This notebook demonstrates how to compute compute contour comparison metrics using PlatiPy.
Import Modules#
[1]:
try:
import platipy
except:
!pip install platipy
import platipy
from pathlib import Path
import pandas as pd
import SimpleITK as sitk
%matplotlib inline
from platipy.imaging.tests.data import download_and_extract_zip_file
from platipy.imaging.label.comparison import (
compute_metric_dsc,
compute_metric_hd,
compute_metric_masd,
compute_volume_metrics,
compute_surface_metrics,
compute_surface_dsc)
from platipy.imaging.visualisation.comparison import contour_comparison
from platipy.imaging.utils.crop import label_to_roi
Download Test Data#
This will download some sample data which was generated using the TCIA LCTSC dataset. The data contains some manual contours as well as auto contours.
Note the contours in this dataset are for demonstration purposes only. No emphasis was placed on the quality of these contours.
[2]:
data_path = Path("./data/contour_comparison_sample")
test_data_zip_url = "https://zenodo.org/record/7519243/files/platipy_contour_comparison_testdata.zip?download=1"
# Only download data if we haven't already downloaded the data previously
if len(list(data_path.glob("*/*.nii.gz"))) == 0:
download_and_extract_zip_file(test_data_zip_url, data_path)
Load data#
Let’s read in the data that we’ve downloaded
[3]:
ct_image = sitk.ReadImage(str(data_path.joinpath("image", "CT.nii.gz")))
structure_names =["ESOPHAGUS", "HEART", "LUNG_L", "LUNG_R", "SPINALCORD"]
manual_structures = {
s: sitk.ReadImage(str(data_path.joinpath("manual", f"{s}.nii.gz"))) for s in structure_names
}
auto_structures = {
s: sitk.ReadImage(str(data_path.joinpath("auto", f"{s}.nii.gz"))) for s in structure_names
}
Compute Metrics (for single structure)#
The following cells demonstrate computing some common metrics between the manual and auto heart contour.
[4]:
heart_dsc = compute_metric_dsc(manual_structures["HEART"], auto_structures["HEART"])
print(f"Dice Similarity Coefficient for Heart is: {heart_dsc:.2f}")
Dice Similarity Coefficient for Heart is: 0.88
[5]:
heart_hd = compute_metric_hd(manual_structures["HEART"], auto_structures["HEART"])
print(f"Hausdorff distance for Heart is: {heart_hd:.2f}mm")
Hausdorff distance for Heart is: 11.92mm
[6]:
heart_masd = compute_metric_masd(manual_structures["HEART"], auto_structures["HEART"])
print(f"Mean Absolute Surface for Heart is: {heart_masd:.2f}mm")
Mean Absolute Surface for Heart is: 4.04mm
[7]:
heart_surface_dsc = compute_surface_dsc(manual_structures["HEART"], auto_structures["HEART"])
print(f"Surface DSC for Heart with a tau of 3mm is: {heart_surface_dsc:.2f}") # 3mm is the default value for tau
heart_surface_dsc = compute_surface_dsc(manual_structures["HEART"], auto_structures["HEART"], tau=5)
print(f"Surface DSC for Heart with a tau of 5mm is: {heart_surface_dsc:.2f}")
Surface DSC for Heart with a tau of 3mm is: 0.52
Surface DSC for Heart with a tau of 5mm is: 0.72
Compute metrics (for multiple structures)#
This example loops over each structure and computes some volume metrics using the compute_volume_metrics helper function. These are tracked in a list and finally converted to a pandas DataFrame.
[8]:
result = []
for structure_name in manual_structures:
volume_dict = compute_volume_metrics(manual_structures[structure_name], auto_structures[structure_name])
structure_dict = {
"structure": structure_name,
**volume_dict,
}
result.append(structure_dict)
df_metrics = pd.DataFrame(result)
df_metrics
[8]:
| structure | DSC | volumeOverlap | fractionOverlap | truePositiveFraction | trueNegativeFraction | falsePositiveFraction | falseNegativeFraction | |
|---|---|---|---|---|---|---|---|---|
| 0 | ESOPHAGUS | 0.430794 | 9.861946 | 0.274530 | 0.357610 | 0.999905 | 0.000095 | 0.642390 |
| 1 | HEART | 0.879741 | 626.272202 | 0.785301 | 0.824823 | 0.999561 | 0.000439 | 0.175177 |
| 2 | LUNG_L | 0.895018 | 953.364372 | 0.809984 | 0.939151 | 0.998134 | 0.001866 | 0.060849 |
| 3 | LUNG_R | 0.905581 | 1330.435753 | 0.827454 | 0.934013 | 0.997875 | 0.002125 | 0.065987 |
| 4 | SPINALCORD | 0.647406 | 33.305168 | 0.478640 | 0.614366 | 0.999825 | 0.000175 | 0.385634 |
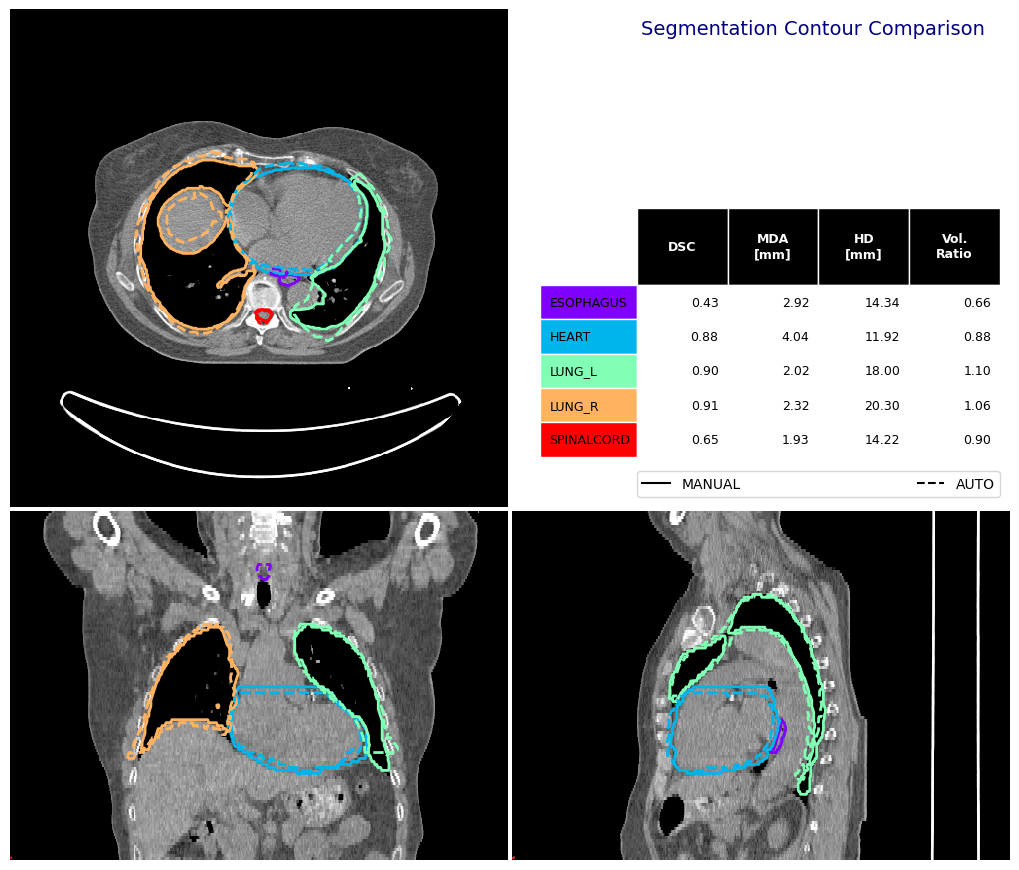
Contour Comparison Function#
You can use the contour_comparison function to prepare a visualisation and return a pandas DataFrame of commonly used metrics.
[9]:
fig, df_mas = contour_comparison(
img = ct_image,
contour_dict_a = manual_structures,
contour_dict_b = auto_structures,
contour_label_a = "MANUAL",
contour_label_b = "AUTO",
structure_for_com = "HEART",
title='Segmentation Contour Comparison',
subtitle='',
subsubtitle='',
)
df_mas
[9]:
| STRUCTURE | DSC | MDA_mm | HD_mm | VOL_MANUAL_cm3 | VOL_AUTO_cm3 | |
|---|---|---|---|---|---|---|
| 0 | ESOPHAGUS | 0.430794 | 2.916249 | 14.344129 | 27.577400 | 18.207550 |
| 0 | HEART | 0.879741 | 4.037049 | 11.917850 | 759.281158 | 664.484024 |
| 0 | LUNG_L | 0.895018 | 2.023395 | 18.000000 | 1015.133858 | 1115.246773 |
| 0 | LUNG_R | 0.905581 | 2.319356 | 20.295816 | 1424.428940 | 1513.873100 |
| 0 | SPINALCORD | 0.647406 | 1.932112 | 14.218965 | 54.210663 | 48.677444 |
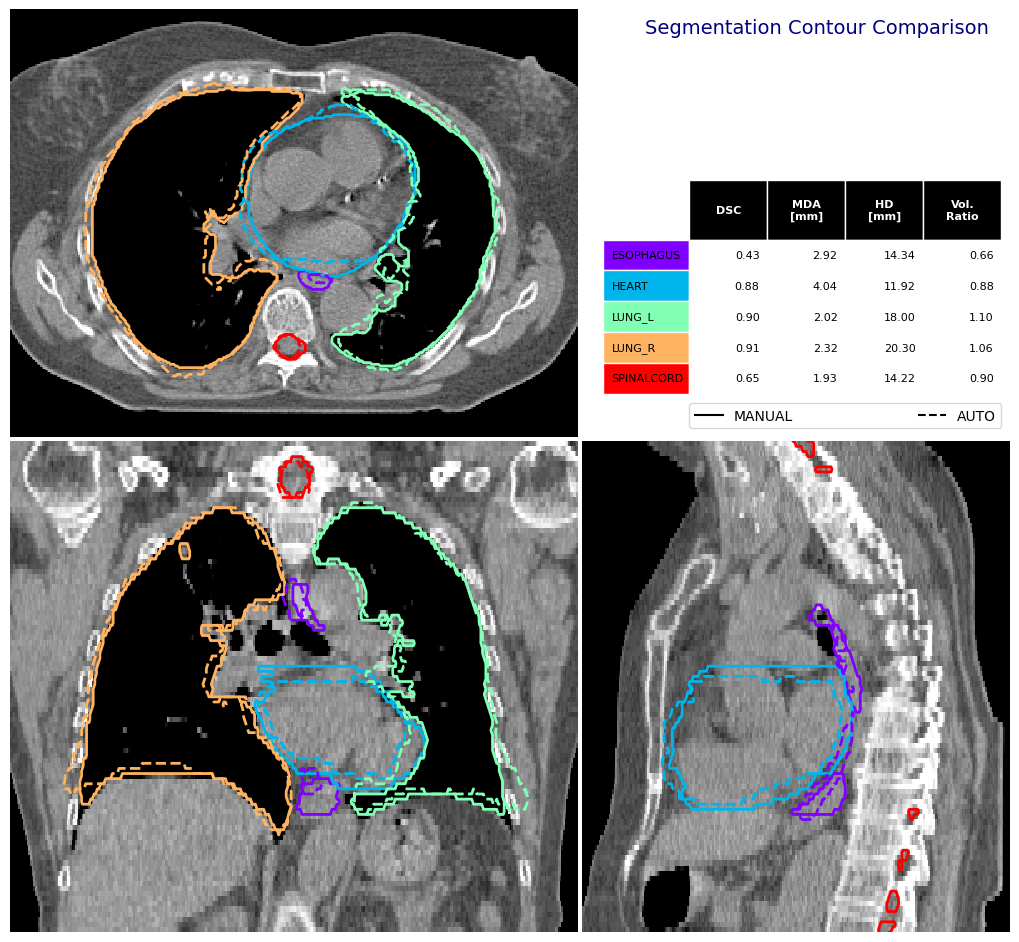
Set limits for visualisation#
You can pass through a dictionary in img_vis_kw to send specific keyword arguments to the ImageVisualiser class. In this example we set the limits of the visualisation to the area of the left and right lungs combined.
[10]:
(sag_size, cor_size, ax_size), (sag_0, cor_0, ax_0) = label_to_roi(
manual_structures["LUNG_L"] | manual_structures["LUNG_R"], expansion_mm=40
)
limits = [ax_0,
ax_0 + ax_size,
cor_0,
cor_0 + cor_size,
sag_0,
sag_0 + sag_size,
]
fig, df_mas = contour_comparison(
img = ct_image,
contour_dict_a = manual_structures,
contour_dict_b = auto_structures,
contour_label_a = "MANUAL",
contour_label_b = "AUTO",
structure_for_com = "ESOPHAGUS",
title='Segmentation Contour Comparison',
subtitle='',
subsubtitle='',
img_vis_kw={"limits": limits},
)
Contour Comparison Reference#
You can find a full list of all contour comparison functions in the PlatiPy documentation.
[ ]: